Cheng Jin
Fudan University
Unified Personalized Reward Model for Vision Generation
Feb 02, 2026Abstract:Recent advancements in multimodal reward models (RMs) have significantly propelled the development of visual generation. Existing frameworks typically adopt Bradley-Terry-style preference modeling or leverage generative VLMs as judges, and subsequently optimize visual generation models via reinforcement learning. However, current RMs suffer from inherent limitations: they often follow a one-size-fits-all paradigm that assumes a monolithic preference distribution or relies on fixed evaluation rubrics. As a result, they are insensitive to content-specific visual cues, leading to systematic misalignment with subjective and context-dependent human preferences. To this end, inspired by human assessment, we propose UnifiedReward-Flex, a unified personalized reward model for vision generation that couples reward modeling with flexible and context-adaptive reasoning. Specifically, given a prompt and the generated visual content, it first interprets the semantic intent and grounds on visual evidence, then dynamically constructs a hierarchical assessment by instantiating fine-grained criteria under both predefined and self-generated high-level dimensions. Our training pipeline follows a two-stage process: (1) we first distill structured, high-quality reasoning traces from advanced closed-source VLMs to bootstrap SFT, equipping the model with flexible and context-adaptive reasoning behaviors; (2) we then perform direct preference optimization (DPO) on carefully curated preference pairs to further strengthen reasoning fidelity and discriminative alignment. To validate the effectiveness, we integrate UnifiedReward-Flex into the GRPO framework for image and video synthesis, and extensive results demonstrate its superiority.
Investigating Data Pruning for Pretraining Biological Foundation Models at Scale
Dec 15, 2025Abstract:Biological foundation models (BioFMs), pretrained on large-scale biological sequences, have recently shown strong potential in providing meaningful representations for diverse downstream bioinformatics tasks. However, such models often rely on millions to billions of training sequences and billions of parameters, resulting in prohibitive computational costs and significant barriers to reproducibility and accessibility, particularly for academic labs. To address these challenges, we investigate the feasibility of data pruning for BioFM pretraining and propose a post-hoc influence-guided data pruning framework tailored to biological domains. Our approach introduces a subset-based self-influence formulation that enables efficient estimation of sample importance at low computational cost, and builds upon it two simple yet effective selection strategies, namely Top-k Influence (Top I) and Coverage-Centric Influence (CCI). We empirically validate our method on two representative BioFMs, RNA-FM and ESM-C. For RNA, our framework consistently outperforms random selection baselines under an extreme pruning rate of over 99 percent, demonstrating its effectiveness. Furthermore, we show the generalizability of our framework on protein-related tasks using ESM-C. In particular, our coreset even outperforms random subsets that are ten times larger in both RNA and protein settings, revealing substantial redundancy in biological sequence datasets. These findings underscore the potential of influence-guided data pruning to substantially reduce the computational cost of BioFM pretraining, paving the way for more efficient, accessible, and sustainable biological AI research.
Explore and Establish Synergistic Effects Between Weight Pruning and Coreset Selection in Neural Network Training
Nov 17, 2025Abstract:Modern deep neural networks rely heavily on massive model weights and training samples, incurring substantial computational costs. Weight pruning and coreset selection are two emerging paradigms proposed to improve computational efficiency. In this paper, we first explore the interplay between redundant weights and training samples through a transparent analysis: redundant samples, particularly noisy ones, cause model weights to become unnecessarily overtuned to fit them, complicating the identification of irrelevant weights during pruning; conversely, irrelevant weights tend to overfit noisy data, undermining coreset selection effectiveness. To further investigate and harness this interplay in deep learning, we develop a Simultaneous Weight and Sample Tailoring mechanism (SWaST) that alternately performs weight pruning and coreset selection to establish a synergistic effect in training. During this investigation, we observe that when simultaneously removing a large number of weights and samples, a phenomenon we term critical double-loss can occur, where important weights and their supportive samples are mistakenly eliminated at the same time, leading to model instability and nearly irreversible degradation that cannot be recovered in subsequent training. Unlike classic machine learning models, this issue can arise in deep learning due to the lack of theoretical guarantees on the correctness of weight pruning and coreset selection, which explains why these paradigms are often developed independently. We mitigate this by integrating a state preservation mechanism into SWaST, enabling stable joint optimization. Extensive experiments reveal a strong synergy between pruning and coreset selection across varying prune rates and coreset sizes, delivering accuracy boosts of up to 17.83% alongside 10% to 90% FLOPs reductions.
Advanced Black-Box Tuning of Large Language Models with Limited API Calls
Nov 17, 2025



Abstract:Black-box tuning is an emerging paradigm for adapting large language models (LLMs) to better achieve desired behaviors, particularly when direct access to model parameters is unavailable. Current strategies, however, often present a dilemma of suboptimal extremes: either separately train a small proxy model and then use it to shift the predictions of the foundation model, offering notable efficiency but often yielding limited improvement; or making API calls in each tuning iteration to the foundation model, which entails prohibitive computational costs. Therefore, we propose a novel advanced black-box tuning method for LLMs with limited API calls. Our core strategy involves training a Gaussian Process (GP) surrogate model with "LogitMap Pairs" derived from querying the foundation model on a minimal but highly informative training subset. This surrogate can approximate the outputs of the foundation model to guide the training of the proxy model, thereby effectively reducing the need for direct queries to the foundation model. Extensive experiments verify that our approach elevates pre-trained language model accuracy from 55.92% to 86.85%, reducing the frequency of API queries to merely 1.38%. This significantly outperforms offline approaches that operate entirely without API access. Notably, our method also achieves comparable or superior accuracy to query-intensive approaches, while significantly reducing API costs. This offers a robust and high-efficiency paradigm for language model adaptation.
Stage-wise Dynamics of Classifier-Free Guidance in Diffusion Models
Sep 26, 2025Abstract:Classifier-Free Guidance (CFG) is widely used to improve conditional fidelity in diffusion models, but its impact on sampling dynamics remains poorly understood. Prior studies, often restricted to unimodal conditional distributions or simplified cases, provide only a partial picture. We analyze CFG under multimodal conditionals and show that the sampling process unfolds in three successive stages. In the Direction Shift stage, guidance accelerates movement toward the weighted mean, introducing initialization bias and norm growth. In the Mode Separation stage, local dynamics remain largely neutral, but the inherited bias suppresses weaker modes, reducing global diversity. In the Concentration stage, guidance amplifies within-mode contraction, diminishing fine-grained variability. This unified view explains a widely observed phenomenon: stronger guidance improves semantic alignment but inevitably reduces diversity. Experiments support these predictions, showing that early strong guidance erodes global diversity, while late strong guidance suppresses fine-grained variation. Moreover, our theory naturally suggests a time-varying guidance schedule, and empirical results confirm that it consistently improves both quality and diversity.
ReTrack: Data Unlearning in Diffusion Models through Redirecting the Denoising Trajectory
Sep 16, 2025Abstract:Diffusion models excel at generating high-quality, diverse images but suffer from training data memorization, raising critical privacy and safety concerns. Data unlearning has emerged to mitigate this issue by removing the influence of specific data without retraining from scratch. We propose ReTrack, a fast and effective data unlearning method for diffusion models. ReTrack employs importance sampling to construct a more efficient fine-tuning loss, which we approximate by retaining only dominant terms. This yields an interpretable objective that redirects denoising trajectories toward the $k$-nearest neighbors, enabling efficient unlearning while preserving generative quality. Experiments on MNIST T-Shirt, CelebA-HQ, CIFAR-10, and Stable Diffusion show that ReTrack achieves state-of-the-art performance, striking the best trade-off between unlearning strength and generation quality preservation.
Pref-GRPO: Pairwise Preference Reward-based GRPO for Stable Text-to-Image Reinforcement Learning
Aug 28, 2025Abstract:Recent advancements highlight the importance of GRPO-based reinforcement learning methods and benchmarking in enhancing text-to-image (T2I) generation. However, current methods using pointwise reward models (RM) for scoring generated images are susceptible to reward hacking. We reveal that this happens when minimal score differences between images are amplified after normalization, creating illusory advantages that drive the model to over-optimize for trivial gains, ultimately destabilizing the image generation process. To address this, we propose Pref-GRPO, a pairwise preference reward-based GRPO method that shifts the optimization objective from score maximization to preference fitting, ensuring more stable training. In Pref-GRPO, images are pairwise compared within each group using preference RM, and the win rate is used as the reward signal. Extensive experiments demonstrate that PREF-GRPO differentiates subtle image quality differences, providing more stable advantages and mitigating reward hacking. Additionally, existing T2I benchmarks are limited by coarse evaluation criteria, hindering comprehensive model assessment. To solve this, we introduce UniGenBench, a unified T2I benchmark comprising 600 prompts across 5 main themes and 20 subthemes. It evaluates semantic consistency through 10 primary and 27 sub-criteria, leveraging MLLM for benchmark construction and evaluation. Our benchmarks uncover the strengths and weaknesses of both open and closed-source T2I models and validate the effectiveness of Pref-GRPO.
A Versatile Pathology Co-pilot via Reasoning Enhanced Multimodal Large Language Model
Jul 23, 2025



Abstract:Multimodal large language models (MLLMs) have emerged as powerful tools for computational pathology, offering unprecedented opportunities to integrate pathological images with language context for comprehensive diagnostic analysis. These models hold particular promise for automating complex tasks that traditionally require expert interpretation of pathologists. However, current MLLM approaches in pathology demonstrate significantly constrained reasoning capabilities, primarily due to their reliance on expensive chain-of-thought annotations. Additionally, existing methods remain limited to simplex application of visual question answering (VQA) at region-of-interest (ROI) level, failing to address the full spectrum of diagnostic needs such as ROI classification, detection, segmentation, whole-slide-image (WSI) classification and VQA in clinical practice. In this study, we present SmartPath-R1, a versatile MLLM capable of simultaneously addressing both ROI-level and WSI-level tasks while demonstrating robust pathological reasoning capability. Our framework combines scale-dependent supervised fine-tuning and task-aware reinforcement fine-tuning, which circumvents the requirement for chain-of-thought supervision by leveraging the intrinsic knowledge within MLLM. Furthermore, SmartPath-R1 integrates multiscale and multitask analysis through a mixture-of-experts mechanism, enabling dynamic processing for diverse tasks. We curate a large-scale dataset comprising 2.3M ROI samples and 188K WSI samples for training and evaluation. Extensive experiments across 72 tasks validate the effectiveness and superiority of the proposed approach. This work represents a significant step toward developing versatile, reasoning-enhanced AI systems for precision pathology.
Genome-Anchored Foundation Model Embeddings Improve Molecular Prediction from Histology Images
Jun 24, 2025

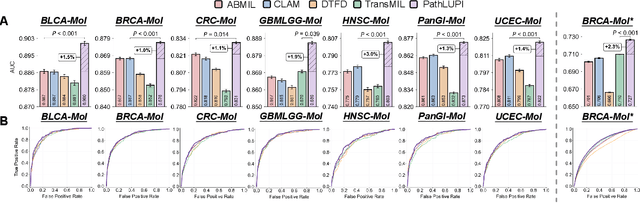

Abstract:Precision oncology requires accurate molecular insights, yet obtaining these directly from genomics is costly and time-consuming for broad clinical use. Predicting complex molecular features and patient prognosis directly from routine whole-slide images (WSI) remains a major challenge for current deep learning methods. Here we introduce PathLUPI, which uses transcriptomic privileged information during training to extract genome-anchored histological embeddings, enabling effective molecular prediction using only WSIs at inference. Through extensive evaluation across 49 molecular oncology tasks using 11,257 cases among 20 cohorts, PathLUPI demonstrated superior performance compared to conventional methods trained solely on WSIs. Crucially, it achieves AUC $\geq$ 0.80 in 14 of the biomarker prediction and molecular subtyping tasks and C-index $\geq$ 0.70 in survival cohorts of 5 major cancer types. Moreover, PathLUPI embeddings reveal distinct cellular morphological signatures associated with specific genotypes and related biological pathways within WSIs. By effectively encoding molecular context to refine WSI representations, PathLUPI overcomes a key limitation of existing models and offers a novel strategy to bridge molecular insights with routine pathology workflows for wider clinical application.
PathBench: A comprehensive comparison benchmark for pathology foundation models towards precision oncology
May 26, 2025



Abstract:The emergence of pathology foundation models has revolutionized computational histopathology, enabling highly accurate, generalized whole-slide image analysis for improved cancer diagnosis, and prognosis assessment. While these models show remarkable potential across cancer diagnostics and prognostics, their clinical translation faces critical challenges including variability in optimal model across cancer types, potential data leakage in evaluation, and lack of standardized benchmarks. Without rigorous, unbiased evaluation, even the most advanced PFMs risk remaining confined to research settings, delaying their life-saving applications. Existing benchmarking efforts remain limited by narrow cancer-type focus, potential pretraining data overlaps, or incomplete task coverage. We present PathBench, the first comprehensive benchmark addressing these gaps through: multi-center in-hourse datasets spanning common cancers with rigorous leakage prevention, evaluation across the full clinical spectrum from diagnosis to prognosis, and an automated leaderboard system for continuous model assessment. Our framework incorporates large-scale data, enabling objective comparison of PFMs while reflecting real-world clinical complexity. All evaluation data comes from private medical providers, with strict exclusion of any pretraining usage to avoid data leakage risks. We have collected 15,888 WSIs from 8,549 patients across 10 hospitals, encompassing over 64 diagnosis and prognosis tasks. Currently, our evaluation of 19 PFMs shows that Virchow2 and H-Optimus-1 are the most effective models overall. This work provides researchers with a robust platform for model development and offers clinicians actionable insights into PFM performance across diverse clinical scenarios, ultimately accelerating the translation of these transformative technologies into routine pathology practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge